Selected Research
Xavi Loinaz, Johnathan Dagan, Jean-Baptiste Alberge, Antonia Kowalewski, Mendy Miller, Julian Hess, Esther Rheinbay, Chip Stewart, Gad Getz
Preprint available on bioRxiv; in preparation for initial submission to Nature Methods
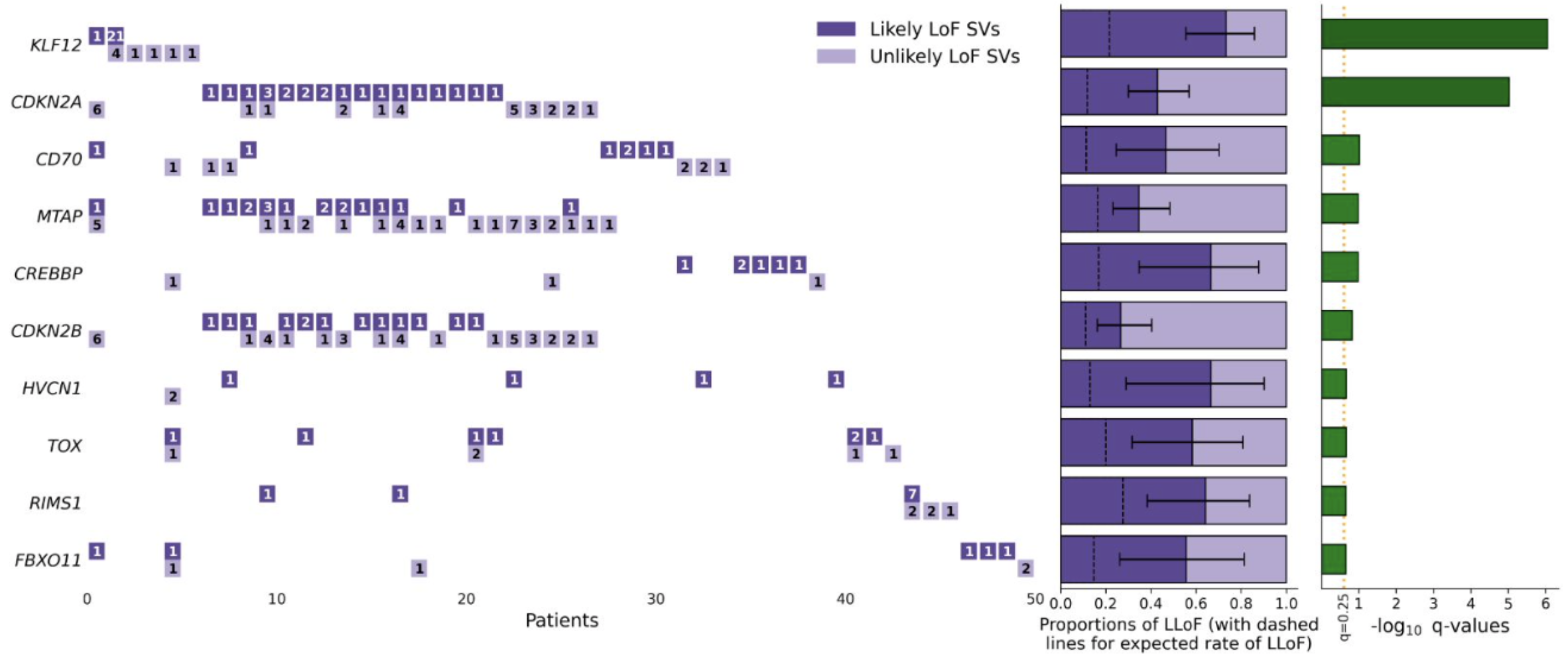 SVelfie (Structural Variants enriched with likely functional/impactful events, pronounced "SVEL-fee") is a statistical method to infer structural variant (SV) driver genes in cancer genome cohorts by analyzing the enrichment of likely functional effects based off breakpoint arrangements. This is an alternative approach to traditional approaches focusing on SV breakpoint frequencies, and it can potentially be used to complement such traditional methods. SVelfie recapitulates known drivers and yields novel candidates, and was applied in a Nature Genetics publication (see below).
SVelfie (Structural Variants enriched with likely functional/impactful events, pronounced "SVEL-fee") is a statistical method to infer structural variant (SV) driver genes in cancer genome cohorts by analyzing the enrichment of likely functional effects based off breakpoint arrangements. This is an alternative approach to traditional approaches focusing on SV breakpoint frequencies, and it can potentially be used to complement such traditional methods. SVelfie recapitulates known drivers and yields novel candidates, and was applied in a Nature Genetics publication (see below).
Jean-Baptiste Alberge*, Ankit K. Dutta*, Andrea Poletti*, Tim H. H. Coorens, Elizabeth D. Lightbody, Rosa Toenges, Xavi Loinaz, Sofia Wallin, Andrew Dunford, Oliver Priebe, Johnathan Dagan, Cody J. Boehner, Erica Horowitz, Nang K. Su, Hadley Barr, Laura Hevenor, Katherine Towle, Rashmika Beesam, Jenna B. Beckwith, Jacqueline Perry, David M. Cordas dos Santos, Luca Bertamini, Patricia T. Greipp, Kirsten Kübler, Peter F. Arndt, Carolina Terragna, Elena Zamagni, Eileen M. Boyle, Kwee Yong, Gareth Morgan, Brian A. Walker, Meletios Athanasios Dimopoulos, Efstathios Kastritis, Julian Hess, Romanos Sklavenitis-Pistofidis, Chip Stewart, Gad Getz**, Irene M. Ghobrial**
Published in Nature Genetics, May 2025
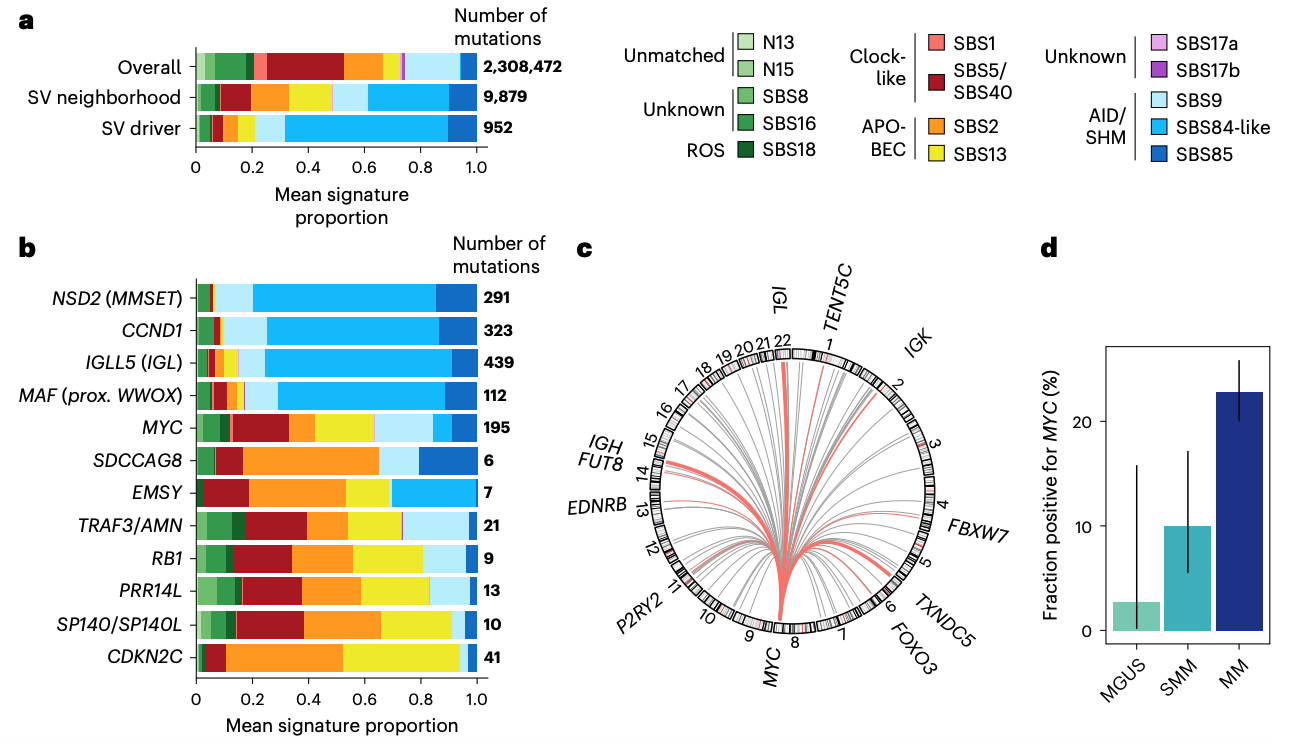 This study presents a comprehensive genomic analysis of multiple myeloma (MM) and its precursor stages, identifying candidate drivers from mutations, copy number alterations, and structural variants from 1,030 patients. Using these findings, an “MM-like” score was developed that predicts disease progression, risk classification, and subclonal dynamics, offering insights to improve early intervention and monitoring strategies. The method I developed, SVelfie (see one above) was used to infer structural variant drivers integrated into this MM-like score, and we also found specific mutational aetiologies associated with such SV candidate drivers (see above graphic).
This study presents a comprehensive genomic analysis of multiple myeloma (MM) and its precursor stages, identifying candidate drivers from mutations, copy number alterations, and structural variants from 1,030 patients. Using these findings, an “MM-like” score was developed that predicts disease progression, risk classification, and subclonal dynamics, offering insights to improve early intervention and monitoring strategies. The method I developed, SVelfie (see one above) was used to infer structural variant drivers integrated into this MM-like score, and we also found specific mutational aetiologies associated with such SV candidate drivers (see above graphic).
Kyle Ellrott*, Christopher K. Wong*, Christina Yau*, Mauro A. A. Castro*, Jordan A. Lee*, Brian J. Karlberg*, Jasleen K. Grewal*, Vincenzo Lagani*, Bahar Tercan*, Verena Friedl, Toshinori Hinoue, Vladislav Uzunangelov, Lindsay Westlake, Xavi Loinaz, Ina Felau, Peggy I. Wang, Anab Kemal, Samantha J. Caesar-Johnson, Ilya Shmulevich, Alexander J. Lazar, Ioannis Tsamardinos, Katherine A. Hoadley, The Cancer Genome Atlas Analysis Network, A. Gordon Robertson, Theo A. Knijnenburg, Christopher C. Benz, Joshua M. Stuart, Jean C. Zenklusen, Andrew D. Cherniack**, Peter W. Laird**
Published in Cancer Cell, January 2025
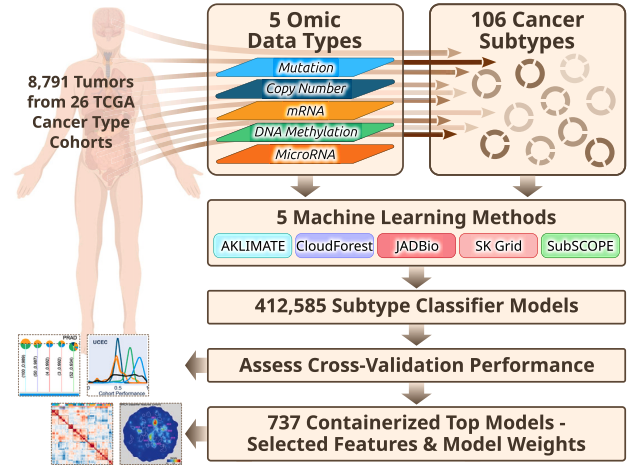 This paper develops machine learning models to classify new cancer samples into predefined TCGA molecular subtypes using multi-omic data from 8,791 tumor samples across 26 cancer cohorts. By validating the models with external datasets and providing containerized versions of the top-performing classifiers, this work attemps to enable more accessible and accurate application of molecular subtyping in clinical and research settings.
This paper develops machine learning models to classify new cancer samples into predefined TCGA molecular subtypes using multi-omic data from 8,791 tumor samples across 26 cancer cohorts. By validating the models with external datasets and providing containerized versions of the top-performing classifiers, this work attemps to enable more accessible and accurate application of molecular subtyping in clinical and research settings.
Jeremy Bigness, Xavi Loinaz, Shalin Patel, Erica Larschan, Ritambhara Singh
Published in the Journal of Computational Biology, May 2022
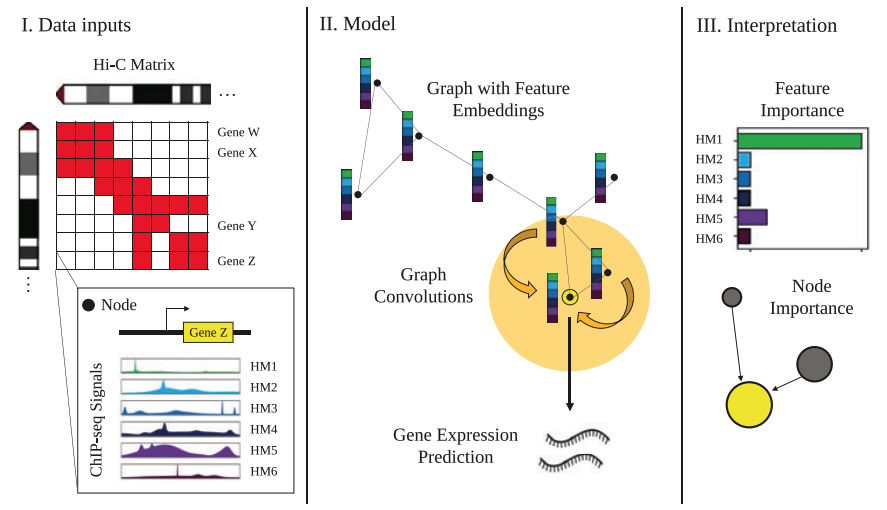 Introduces GC-MERGE, a graph convolutional model that integrates long-range genomic interactions and local regulatory factors (specifically histone modifications) to predict gene expression while capturing the 3D structure of the genome. By applying the model to multiple cell lines, the study demonstrates state-of-the-art predictive performance and interprets the biological mechanisms behind its predictions, offering potential insights into the local and long-range regulatory factors influencing gene expression.
Introduces GC-MERGE, a graph convolutional model that integrates long-range genomic interactions and local regulatory factors (specifically histone modifications) to predict gene expression while capturing the 3D structure of the genome. By applying the model to multiple cell lines, the study demonstrates state-of-the-art predictive performance and interprets the biological mechanisms behind its predictions, offering potential insights into the local and long-range regulatory factors influencing gene expression.
Xavi Loinaz (advised by Ritambhara Singh)
Brown University Honors Senior Thesis, May 2021
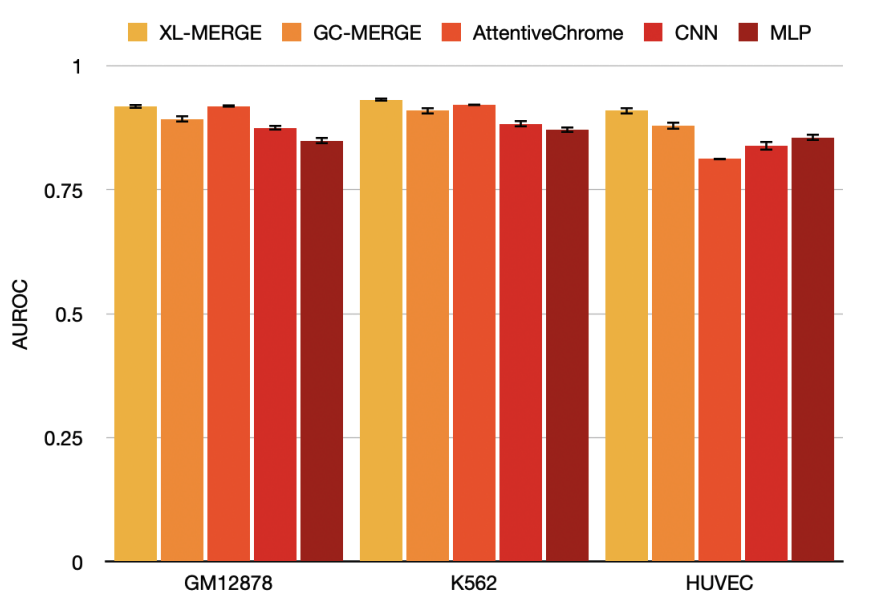 Introduces XL-MERGE, a modified version of GC-MERGE with three different mechanisms for improvement, outperforming GC-MERGE for all three tested cell lines in terms of AUROC.
Introduces XL-MERGE, a modified version of GC-MERGE with three different mechanisms for improvement, outperforming GC-MERGE for all three tested cell lines in terms of AUROC.
Selected Other Projects
Characterization of somatic events for diffuse large B-cell lymphoma in the Clinical Trial Sequencing Project
As the Getz Lab's lead analyst of the NIH-funded Clinical Trial Sequencing Project (CTSP) on diffuse large B-cell lymphoma, I fixed pipelines and analyzed somatic mutations, indels, copy number alterations, structural variants, and mutational signatures in 138 diffuse large B-cell lymphoma (DLBCL) samples. Had poster presentation for 2021 Broad Retreat Scientific Retreat; content is unable to be posted due to consortium rules. Project is progressing towards a manuscript.
Characterization and concordance analysis for cell lines of the Human Cancer Models Initiative
Acted as the Getz Lab's lead analyst for the NIH-funded Human Cancer Models Initiative to develop next-generation cancer models that better represent the diversity and complexity of human cancers. I focused on analysis relating to structural variation and quality control, as well as concordance of mutational signatures between tumor samples and corresponding models. Associated manuscripts under review.
Getz Lab's Structural Variant-Calling Pipeline
I rewrote and maintain the tumor structural variant-calling pipeline of the Getz Lab (50+-person lab). I also implemented a scatter/gather parallelization of the SV-calling tool SvABA, reducing SvABA’s runtime by a factor of ~10, and additionally have patched several bugs in the pipeline.
For spring semester of my junior year in college, I had an independent study research project where I developed a proof-of-concept system for data synchronization between video and various electrophysiological sensors for studies in Prof. David Borton's lab at Brown. I read in and decoded LTC signals from Timecode Systems devices on an Arduino and wrote software to test synchronization. I also resolved a problem in drift between GoPro and behavioral sensor data that had affected one of the lab's projects and had been unresolved for about a year.
I published a brief article on FanGraphs' community research blog on evaluation of different predictive pitching metrics.
Back in high school, I conceptualized and led the development and maintenance of an official school-sanctioned iOS app with useful features for my high school's community. It eventually got over 5,000 downloads and was regularly used by students, parents, and faculty. This was my first formative experience in seeing how immediate of an impact computational technologies I could develop could have on the people around me!
[Press]
Mentorship and Outreach
I love mentoring students and building both their enthusiasm and skills in research and academics! I have mentored three undergraduate students in Getz Lab, and used to be a classroom leader for the after-school volunteering program Buddies4Math.
 Me with my undergraduate intern, Johnathan, at the Getz Lab's end-of-summer intern poster showcase!
Me with my undergraduate intern, Johnathan, at the Getz Lab's end-of-summer intern poster showcase!
Additionally, I was part of the steering committee for RATalks, a group at the Broad Institute that hosts talks to the greater community. For this, I conceptualized and led planning of our very first alumni RATalk which sought to disseminate career and life perspective to the early-researcher community by a former associate director of the Broad Institute, increasing the availability of mentorship to junior researchers!
Reading and Writing
I love to read and average around 30 books per year. What I love most about reading is how it enables better understanding of those around us and their perspectives. I think this is especially important in research, where this can help me to be a better collaborator and also to create a more welcoming environment to others from diverse backgrounds. Some books that have inspired me:
- Free Food for Millionaires by Min Jin Lee
- Everything I Never Told You by Celeste Ng
- Little Failure by Gary Shteyngart
- Deep Work: Rules for Focused Success in a Distracted World by Cal Newport
- Miracle Creek by Angie Kim
- How to Know a Person: The Art of Seeing Others Deeply and Being Deeply Seen by David Brooks
- Hunger: A Memoir of (My) Body by Roxane Gay
- Born a Crime: Stories From a South African Childhood by Trevor Noah
- Daring Greatly: How the Courage to Be Vulnerable Transforms the Way We Live, Love, Parent, and Lead by Brené Brown
Due in part to the perspectives I gained from my reading, I also wrote an essay about how to increase diversity in computer science-related disciplines, which incidentally helped earn me $159,000 of funding for my PhD via the NSF's CSGrad4US fellowship.
This website's design was based on a template by
Shouvik Mani.

 SVelfie (Structural Variants enriched with likely functional/impactful events, pronounced "SVEL-fee") is a statistical method to infer structural variant (SV) driver genes in cancer genome cohorts by analyzing the enrichment of likely functional effects based off breakpoint arrangements. This is an alternative approach to traditional approaches focusing on SV breakpoint frequencies, and it can potentially be used to complement such traditional methods. SVelfie recapitulates known drivers and yields novel candidates, and was applied in a Nature Genetics publication (see below).
SVelfie (Structural Variants enriched with likely functional/impactful events, pronounced "SVEL-fee") is a statistical method to infer structural variant (SV) driver genes in cancer genome cohorts by analyzing the enrichment of likely functional effects based off breakpoint arrangements. This is an alternative approach to traditional approaches focusing on SV breakpoint frequencies, and it can potentially be used to complement such traditional methods. SVelfie recapitulates known drivers and yields novel candidates, and was applied in a Nature Genetics publication (see below).
 This study presents a comprehensive genomic analysis of multiple myeloma (MM) and its precursor stages, identifying candidate drivers from mutations, copy number alterations, and structural variants from 1,030 patients. Using these findings, an “MM-like” score was developed that predicts disease progression, risk classification, and subclonal dynamics, offering insights to improve early intervention and monitoring strategies. The method I developed, SVelfie (see one above) was used to infer structural variant drivers integrated into this MM-like score, and we also found specific mutational aetiologies associated with such SV candidate drivers (see above graphic).
This study presents a comprehensive genomic analysis of multiple myeloma (MM) and its precursor stages, identifying candidate drivers from mutations, copy number alterations, and structural variants from 1,030 patients. Using these findings, an “MM-like” score was developed that predicts disease progression, risk classification, and subclonal dynamics, offering insights to improve early intervention and monitoring strategies. The method I developed, SVelfie (see one above) was used to infer structural variant drivers integrated into this MM-like score, and we also found specific mutational aetiologies associated with such SV candidate drivers (see above graphic).
 This paper develops machine learning models to classify new cancer samples into predefined TCGA molecular subtypes using multi-omic data from 8,791 tumor samples across 26 cancer cohorts. By validating the models with external datasets and providing containerized versions of the top-performing classifiers, this work attemps to enable more accessible and accurate application of molecular subtyping in clinical and research settings.
This paper develops machine learning models to classify new cancer samples into predefined TCGA molecular subtypes using multi-omic data from 8,791 tumor samples across 26 cancer cohorts. By validating the models with external datasets and providing containerized versions of the top-performing classifiers, this work attemps to enable more accessible and accurate application of molecular subtyping in clinical and research settings.
 Introduces GC-MERGE, a graph convolutional model that integrates long-range genomic interactions and local regulatory factors (specifically histone modifications) to predict gene expression while capturing the 3D structure of the genome. By applying the model to multiple cell lines, the study demonstrates state-of-the-art predictive performance and interprets the biological mechanisms behind its predictions, offering potential insights into the local and long-range regulatory factors influencing gene expression.
Introduces GC-MERGE, a graph convolutional model that integrates long-range genomic interactions and local regulatory factors (specifically histone modifications) to predict gene expression while capturing the 3D structure of the genome. By applying the model to multiple cell lines, the study demonstrates state-of-the-art predictive performance and interprets the biological mechanisms behind its predictions, offering potential insights into the local and long-range regulatory factors influencing gene expression.
 Introduces XL-MERGE, a modified version of GC-MERGE with three different mechanisms for improvement, outperforming GC-MERGE for all three tested cell lines in terms of AUROC.
Introduces XL-MERGE, a modified version of GC-MERGE with three different mechanisms for improvement, outperforming GC-MERGE for all three tested cell lines in terms of AUROC.
